mCAFE: Microbial community analysis and functional evaluation in soils
The mCAFEs project is a proposed 10-year Department of Energy (DOE) Science Focus Area (SFA) project with the goal of developing the foundational tools and molecular ecosystems biology-based understanding required to predict, manipulate, and design rhizosphere microbial communities. The project has two primary research foci: i) Tool Development and ii) Tool-Enabled Systems Biology in Rhizosphere Microbial Communities.
i) Tool Development: As part of our tool development we are attempting to pioneer microbial community editing tools for dissecting the function of genes, pathways, and microbes within complex communities. This includes technologies for detection of transformation efficiency, DNA delivery, targeted CRISPR-Cas based editing, and microbial ablation. While this represents a significant challenge, microorganisms do not naturally grow as isolates, and the development of genetic technologies to manipulate microbes in the context of their natural communities will be transformative for understanding microbial community dynamics.
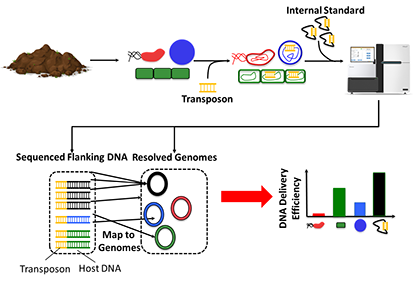
a) ET-Seq: Editing organisms in a complex microbiome requires knowledge of which members are accessible to nucleic acid delivery and editing. Environmental Transformation Sequencing (ET-Seq) is a technology developed to assess the ability of individual species within a microbial community to acquire and integrate exogenous DNA without the need to isolate any individual members. In ET-seq, a microbial community is exposed to a randomly integrating mobile genetic and, in the absence of any selection, total community DNA is extracted and sequenced using two protocols. In the first, we enrich and sequence the junctions between the inserted and host DNA to determine insertion location and quantity in each host. In the second protocol, we use low-depth metagenomic sequencing to quantify the abundance of each community member in a sample. Together, these sequencing procedures provide a quantitative measure of transformation efficiency for microbiota members.
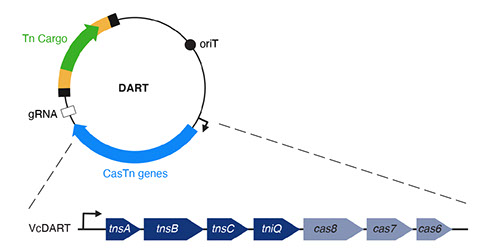
b) DART: Once it is determined that an organism in a microbial community can acquire and integrate exogenous DNA, a method is necessary to target DNA delivered to the community to that specific organism and only that organism. For this we have developed DNA-editing all-in-one RNA-guided CRISPR–Cas transposase (DART) systems for locus-specific insertion of DNA into organisms within a microbial community. The DART system uses an RNA-guided CRISPR–Cas Tn7 transposase to deliver cargo DNA that is between transposon repeats on a plasmid into a specific location of a target genome defined by the guide RNA (gRNA) also encoded on the same plasmid. These systems also include barcodes that make them compatible with ET-Seq to allow for the detection and tracking of uniquely edited cells. Thus, DART can be used seamlessly with ET-seq for rapid assay of the efficacy of CRISPR–Cas-guided transposition into the genome of a target organism in the absence of selectable markers.
c) EcoFAB: . We are developing a fabricated ecosystem (EcoFAB) that can serve as a standardized and reproducible analysis platform for both synthetic consortia and natural microbiomes in simulated and natural environments under controlled conditions. The system is particularly suited for the study of rhizosphere microbiology as it accommodates plant growth, and allows live cell imaging.
ii) Tool Enabled Systems Biology: In this part of the project the tools such as EcoFABs and community editing techniques are combined with a pipeline of large-scale phenotyping, genetics, and metabolomics to greatly improve our understanding of the gene functions of rhizosphere microbes. Using combinations of rhizosphere isolates as well as naturally enriched communities, we are applying full spectrumomics technologies (metagenomics, transcriptomics, and proteomics) to characterize the activities, and interactions of microbial assemblages.



Learn more about the project at https://mcafes.lbl.gov/

